Colloquium discusses complex networks and evolution
In last Wednesday’s colloquium, researcher Roberto Andrade, from the Universidade Federal da Bahia, talked about his latest works involving complex networks for the study of evolution. Andrade and his group propose a new method for organizing species and their evolution, traditionally done through phylogenetic trees based on morphology and molecular characteristics.
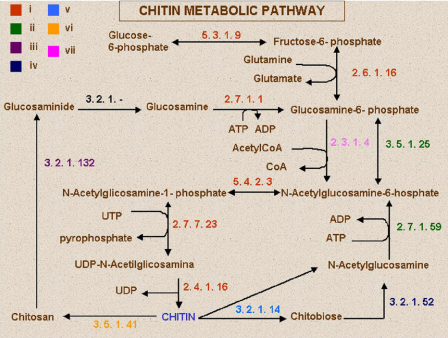
“Our proposal is to classify organisms based on similarities of components involved in the synthesis of basic molecules”, says Andrade. “In our work, we studied fungi based on chitin. Then, in a more ambitious project, we studied the evolutionary origins of the mitochondria”.
The researcher explains that many organisms have their own set of enzymes to obtain a molecule that is found in many different species. Therefore, by studying differences between cell machinery, it’s possible to classify organisms by their genetic proximity.
This situation happens, for instance, with chitin – a molecule that is the main component of fungal cell wall. Different species, however, obtain chitin through different processes. Andrade and his group used the NCBI (National Center for Biotechnology Information) database to compare the molecular mechanisms of various species.
“The classification we reached coincided 100% with other phylogenetic trees”, says Andrade. “However, the method we used to obtain the final classification is faster”.
Origins of the mitochondria
“After this work, we got more ambitious”, says Andrade. “We started working on a project to discover the evolutionary origins of the mitochondria”.
Although many theories attempt to explain the evolution of the mitochondria, the issue still intrigues scientists. For those unfamiliar with biology, mitochondria is an organelle that has its own DNA. Some hypothesis, such as Endosymbiotic Theory, proposes that mitochondria originated when a prokaryotic organism (which has no cell nucleus) started living in symbiosis with a eukaryotic cell (which has cell nucleus).
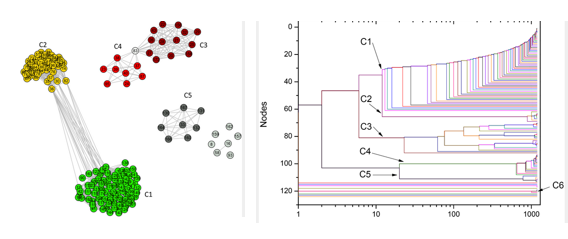
In the left, complex networks shows the proximity of different groups; in the right, the resulting phylogenetic tree
This time, the study was based on the ATP synthesis process – this molecule is considered a cellular fuel, since cells obtain energy by breaking it. Andrade and his group compared the molecular structures that lead to ATP synthesis in bacteria and in mitochondria, to find out which bacteria were most genetically close to the organelle.
“Our main result was that Rickettsiales bacteria are not closely related to mitochondria”, says Andrade. “The work indicated that Rhodobacterales, Rhizobiales and Rhodospirillales are groups closer to the organelle”.
The work was published this year in PLOS and can be found here.